More than 6.5 million Americans live with Alzheimer’s disease – the most common cause of dementia in older adults – according to the Centers for Disease Control and Prevention.
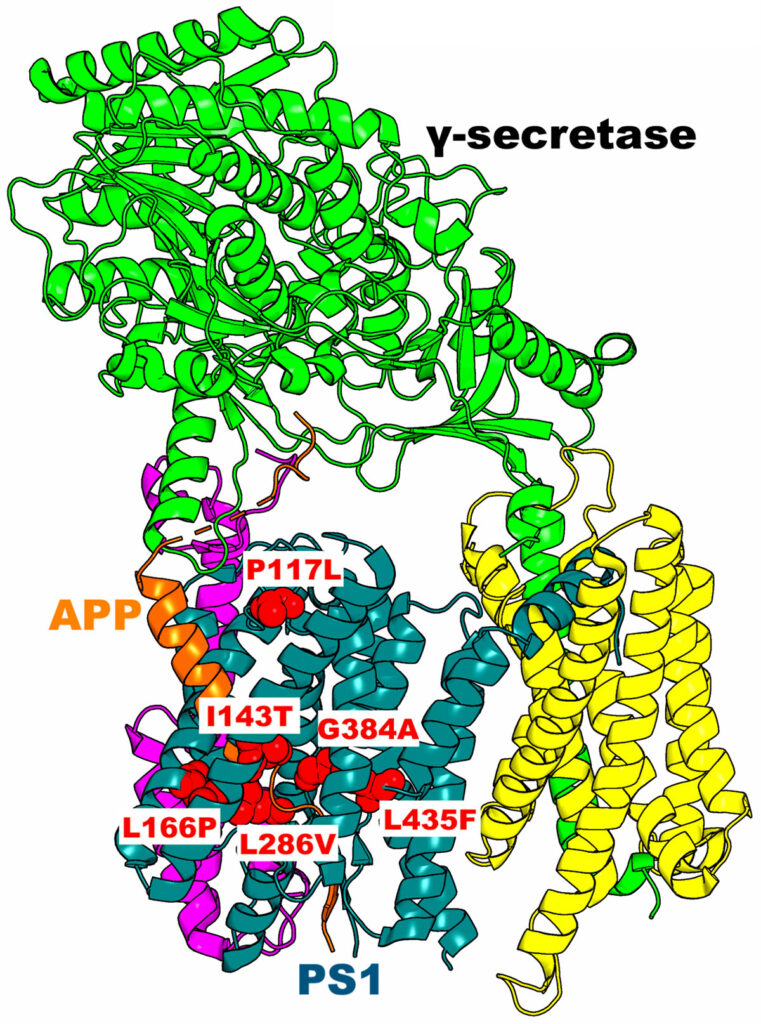
Researchers who study familial Alzheimer’s disease (FAD) – a rare form of Alzheimer’s disease caused by changes in inherited genes – turned to Expanse at the San Diego Supercomputer Center at UC San Diego to conduct computational biology studies on how mutations of a critical protein enzyme called γ-secretase could be treated to better control thought, language and memory.
University of Kansas (KU) graduate students Hung Do, Kushal Koirala and Apurba Bhattarai, led by Associate Professor of Molecular Biosciences Yinglong Miao, used the Expanse supercomputer to build the first dynamic models that show the activation and substrate processing of γ-secretase in the absence and presence of various FAD mutations. The team also worked with KU Professor of Medicinal Chemistry Michael Wolfe and Postdoctoral Researcher Sujan Devkota to compare Wolfe’s lab’s biochemical experimental data with Miao’s team’s computational biology data.
The researchers published their findings in ACS Central Science, Journal of American Chemical Society and more recently in the Nature Communications Biology Journal, reporting that their studies provide a mechanistic basis to guide their rational drug discovery efforts of γ-secretase for treating Alzheimer’s disease. Additionally, their laboratories were able to combine cutting-edge accelerated molecular simulations using Expanse GPUs with highly complementary mass spectrometry and western blot biochemical experiments to investigate the functional mechanisms of substrate cleavage by γ-secretase and the effects of these FAD mutations.
Our primary objectives are to obtain comprehensive understanding of the structural dynamics and functional mechanisms of γ-secretase in the wildtype and disease-causing mutant forms; determine the mechanisms of recognition and processing of various substrates by γ-secretase; and design selective and potent drug molecules of γ-secretase to treat Alzheimer’s disease and other related human diseases. Supercomputers like Expanse allow us to accomplish these goals – one simulation at a time.
Yinglong Miao, KU associate professor of Molecular Biosciences
Wolfe noted that the collaboration between medicinal chemists, such as those in his group, and computational biologists, such as those in Miao’s group, moves scientists closer to treating this devastating disease.
You can read more about this story here (originally published by SDSC, Feb. 23, 2023): Computational Biology Studies Move Researchers Closer to Treating Rare Form of Alzheimer’s Disease.
Project Details
Institution: SDSC (San Diego Supercomputer Center)
University: University of Kansas
Funding Agency: This work was funded by the National Science Foundation (award no. 2121063) and the National Institutes of Health (award no. AG66986). Supercomputing time on Expanse was funded by the NSF’s Extreme Science and Engineering Discovery Environment (allocation no. MCB180049).
Grant Number: See above.
The science story featured here, allocated through August 31, 2022, was enabled through Extreme Science and Engineering Discovery Environment (XSEDE) and supported by National Science Foundation grant number #1548562. Projects allocated September 1, 2022 and beyond are enabled by the ACCESS program, which is supported by National Science Foundation grants #2138259, #2138286, #2138307, #2137603, and #2138296.


