SDSC’s Neha Srini contributed to this piece.
Marine amoeba, a pivotal player in marine ecosystems, serves as both a food source for larger organisms and a habitat for microorganisms. A research team led by Yonas Tekle at Spelman College has been utilizing National Science Foundation (NSF)-funded supercomputers for the past six years to unravel the complexities of Trichosphareium, an enigmatic marine amoeba with a complex life cycle.
Their recent publication, Omics of an Enigmatic Marine Amoeba Uncovers Unprecedented Gene Trafficking from Giant Viruses and Provides Insights into Its Complex Life Cycle in the Microbiology Research Journal, sheds light on their research endeavors.
Professor Tekle, who specializes in Microbial Diversity at Spelman, explained the significance of supercomputers in their research. “Without the supercomputers, most of our computationally extensive genomic work would not be possible, as our local machines have limited power to conduct such detailed studies.”
The research team, which includes colleagues Hanh Tran, Fang Wang, Mandakini Singla and Isimeme Udu, utilizes a range of ACCESS machines, including Bridges-2 at the Pittsburgh Supercomputing Center (PSC) and Jetstream2 at the University of Indiana.
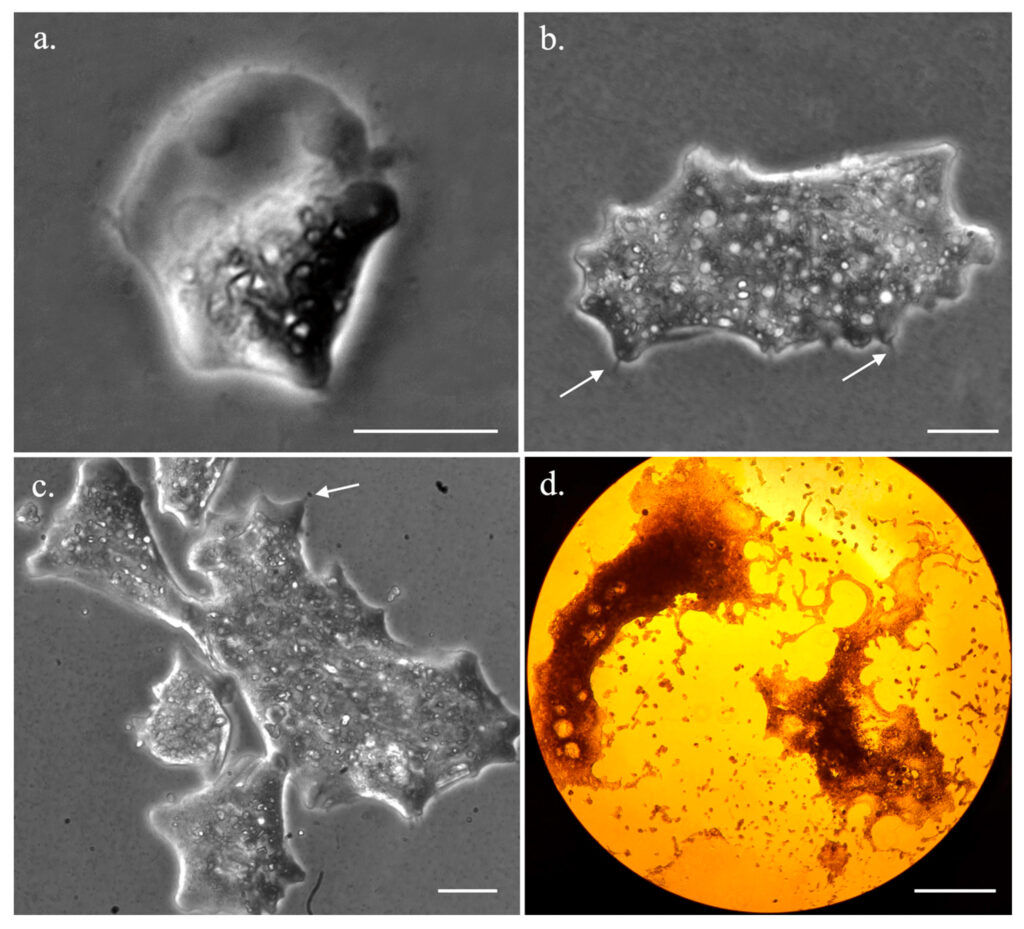
Their studies encompass various aspects of Trichosphareium, ranging from morphology and life cycle to genetic constitution. The team adopts an omics approach – combining genomics, proteomics, transcriptomics and advanced microscopy to observe the life cycle.
Without the supercomputers, most of our computationally extensive genomic work would not be possible, as our local machines have limited power to conduct such detailed studies.
Yonas Tekle, professor, Spelman College
Tekle said that supercomputers are crucial for running simulations to categorize and analyze their data. They explained that their findings have significant implications – particularly in constructing the tree of life with a focus on amoeboid organisms and elucidating cryptic sexual behavior in microbial eukaryotes.
Professor Tekle emphasized the importance of their research for the future.
“There is so much we do not know about microbial eukaryotes that play a crucial role in the environment,” they said. “These microbes – commonly known as protists – are understudied. A comprehensive approach, such as omics studies, will unravel their contribution to the health of our planet and humans.”
Project Details
Resource Provider Institution(s): Pittsburgh Supercomputing Center (PSC), Indiana University
Affiliations: Spelman College
Funding Agency: NSF
Grant or Allocation Number(s): The research was supported by the National Institutes of Health (grant no. 1R15GM116103-02) and the National Science Foundation (grant no. 1831958). Computational work was funded by ACCESS (grant no. TRA180031)
The science story featured here was enabled by the ACCESS program, which is supported by National Science Foundation grants #2138259, #2138286, #2138307, #2137603, and #2138296.


