Many promising antibiotics have been discovered that would potentially outperform the current standard treatment for tuberculosis (TB), which kills 1.5 million people every year. However, determining the best combination of medicines for TB treatment is challenging, expensive and time-consuming. For the past three decades, University of Michigan Professor of Microbiology and Immunology Denise Kirschner has been working on an array of research projects related to TB – including how best to configure appropriate treatments for TB patients using computational modeling.
Kirschner and her team most recently utilized ACCESS allocations on Expanse at the San Diego Supercomputer Center (SDSC) at UC San Diego and Anvil at Purdue University to generate models that showed how TB lesions treated with various combinations of drugs respond to various treatments of combined antibiotics. The results from their study were then compared with results from clinical trials and animal studies.
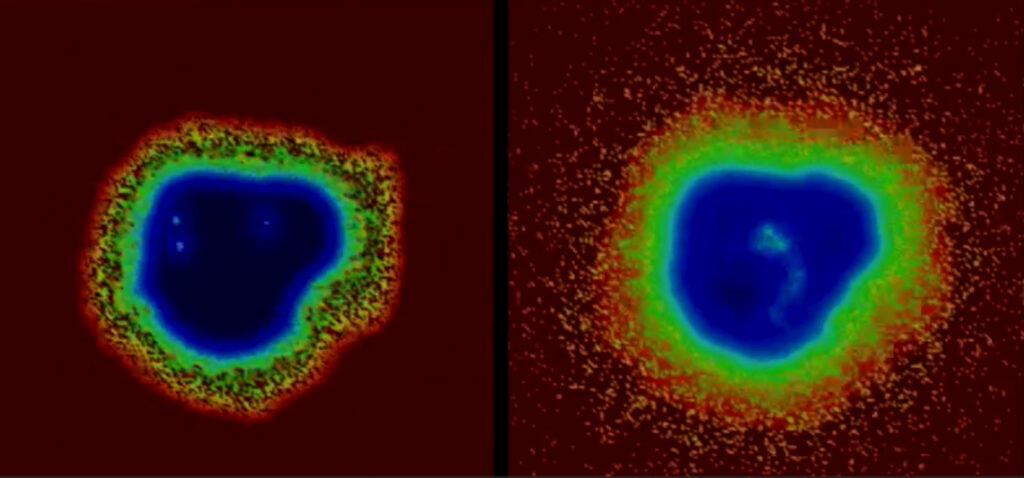
The researchers found that their supercomputer-generated computational models agreed with both the clinical trial and animal study results. They published the study details in CPT: Pharmacometrics & Systems Pharmacology.
“This process is a cheaper and faster alternative to traditional approaches where drugs are tested on humans or animals,” Kirschner said. “Our team’s modeling approach is a vital method of helping physicians narrow down possible drug combinations for TB and could be adapted for cancers that present solid tumors similar to the consistency of TB lesions.”
“ACCESS saved us both time and money. Using local university resources at the scale needed for our research would have been prohibitively expensive while the availability of a large number of cores via ACCESS machines meant that we could run thousands of simulations in parallel.”
–Denise Kirschner, University of Michigan Professor of Microbiology and Immunology
The next step for Kirschner’s lab is to simulate drug combinations that haven’t been tested on humans or animals.
“Our ultimate goal is convincing experimental researchers to test our predicted optimal combinations,” she said. “And, without ACCESS resources it would not be possible to do this research since the time on the other resources we have available would take years.”
Project Details
Resource Provider Institution(s): San Diego Supercomputing Center (SDSC), Rosen Center for Advanced Computing (RCAC)
Affiliations: University of Michigan
Funding Agency: NSF
Grant or Allocation Number(s): MCB140228
The science story featured here was enabled by the U.S. National Science Foundation’s ACCESS program, which is supported by National Science Foundation grants #2138259, #2138286, #2138307, #2137603, and #2138296.